Protein Composition and Structure
Composition
The database contains 37,866 proteins representing 25,770 named loci. For each locus, a largest isoform was selected for compilation of the statistics that follow. These 25,770 proteins have a mean size of 483 amino acids (aa) and a median of 343 aa. A considerable fraction of the proteins in the data set derive from computational predictions. When these are excluded, the mean increases to 575 aa and the median to 431 aa. This smaller set of 18,886 proteins ranges in size from 25 to 33,423 residues. A single protein in this set, 58 aa LUZP6, starts with an isoleucine rather than the usual methionine.In the following table, two methods were used to calculate amino acid usage in the 18,886 selected proteins. In the "By protein" column, compositions were calculated for each of the proteins and then averaged. In the "By sequence" column, the usage is from treating the 18,886 sequences as one long sequence. The latter method is weighted toward the usage in larger proteins. These numbers are not weighted for expression.
Amino acid Usage (%)
By protein By sequence
A alanine 7.214 7.010
C cysteine 2.491 2.284
D aspartate 4.591 4.767
E glutamate 6.839 7.124
F phenylalanine 3.830 3.664
G glycine 6.716 6.577
H histidine 2.592 2.623
I isoleucine 4.378 4.352
K lysine 5.749 5.745
L leucine 10.091 9.964
M methionine 2.284 2.138
N asparagine 3.484 3.603
P proline 6.174 6.285
Q glutamine 4.578 4.751
R arginine 5.804 5.636
S serine 7.944 8.302
T threonine 5.149 5.315
U selenocysteine 0.001 0.000
V valine 6.023 5.980
W tryptophan 1.277 1.207
Y tyrosine 2.793 2.670
There are significant variations from the values above in the usage of many amino acids at the amino termini and carboxyl termini of proteins. These differences may be related to frequent modifications, or other processing and degradation pathways. One example of note is the elevated level of cysteine four positions from the carboxyl terminus, likely reflecting prenylation.
The genome encodes several families of proteins with very unusual amino acid compositions. Many of these are smaller proteins such as the protamines, late cornified envelope proteins, and metallothioneins.
The following table provides some additional examples of individual proteins and gene families where larger proteins have unusual compositions. The numbers given are residues for that amino acid and the total size of the protein. Some predicted proteins have been excluded. The relative fractions vary among the amino acids with the tryptophan-rich proteins being considerably lower than the others. For additional imformation about these proteins, see the sections listed in the right column of the table
Many proteins contain short proline-rich regions. Some proteins, such as certain members of the formin family have very large proline-rich regions that affect the overall composition of the proteins. A similar situation is seen with the leucine-rich repeat proteins.
The small number of proteins containing selenocysteine are described separately (see Selenium Proteins).
Homopolymer segments
Many protein sequences contain long runs of a single amino acid. Notable examples from the largest isoforms in the reference set are presented in the following table (some predicted proteins have been excluded). Proteins often have much larger regions where runs of a single amino acid are broken by one or a few other amino acids. The homopolymer tracts may not be encoded using a single codon for that amino acid. Such variation in codon usage would increase the stability of the DNA sequences that encode the homopolymer tracts. The proteins are described in the sections listed in the right column.
Very large proteins
The following table provides a list of the largest proteins in the reference set. Only one isoform is listed for each. Predicted proteins are not listed. Note also the very large predicted LOC643677 (7081 aa) and HMCN2 (5065 aa).
Many of the proteins listed above contain spectrin-type repeats. Additional large proteins are listed with that family. Larger proteins often contain repeating domains such as those first identified in epidermal growth factor and fibronectin.
Protein modifications
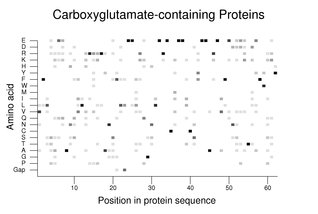
Peptide processing and posttranslational modifications of proteins are presented in detail in the chapters on Proteases and Translation and Protein Modification. The presence of large gene families for proteins that are the substrates for such modifications can be helpful in identifying sequences important for these functions.Proteins with the γ-carboxyglutamate modification are described in the section on coagulation. The following figure shows the amino acid usage (darker being more conserved) in a partial alignment of 11 of these proteins (see Notes and References). Note the completely conserved glutamate residues near the center of the alignments. Interpretation of such alignments can be complex. In this case, a number of these proteins are also processed by cleavage amino-terminal to the relatively conserved alanine at position 18 in the figure.
Another example of shared sequences around the location of a modified amino acid is seen at the active site of sulfatases. In these enzymes, a cysteine is converted to formylglycine.
Notes and references
Many references and other information for individual genes can be found in the RefSeq entries linked via the pages for the proteins mentioned in this section. A table of these entries (with the corresponding gene identifiers) and a collection of their sequences also are available.The tables in this section were constructed using the human RefSeq proteins set available at the time release 37.1 of the human reference genome sequence became available. There are some differences in this protein set and the genes annotated onto the reference genome.
The RefSeq proteins are associated with specific transcripts and there are often multiple transcripts for a given gene that may produce distinct or identical protein products. As explained in this section, this protein set was reduced by eliminating gene predictions and then choosing a single largest isoform for each gene. Also, only protein sequences derived from the reference mitochondrial genome were retained.
To produce the figure on carboxyglutamate-containing proteins, amino acids 24-85 from PROZ were used in searches to produce the alignments. The proteins used are those listed in the example in the section on coagulation except for PRRG2. MGP and BGLAP were also omitted.
See also the additional reading for this chapter.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.