
| Name: GAST | Sequence: fasta or formatted (101aa) | NCBI GI: 4503923 | |
|
Description: gastrin preproprotein
|
Referenced in: Stomach, Small Intestine, and Colon
| ||
|
Composition:
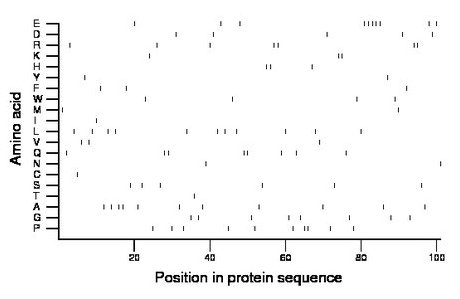
Amino acid Percentage Count Longest homopolymer A alanine 10.9 11 2 C cysteine 1.0 1 1 D aspartate 5.0 5 1 E glutamate 9.9 10 5 F phenylalanine 3.0 3 1 G glycine 7.9 8 1 H histidine 3.0 3 2 I isoleucine 1.0 1 1 K lysine 3.0 3 2 L leucine 10.9 11 1 M methionine 2.0 2 1 N asparagine 2.0 2 1 P proline 9.9 10 2 Q glutamine 7.9 8 2 R arginine 6.9 7 2 S serine 5.9 6 1 T threonine 1.0 1 1 V valine 3.0 3 1 W tryptophan 4.0 4 1 Y tyrosine 2.0 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 gastrin preproprotein CCK 0.052 cholecystokinin preproprotein KRBA1 0.031 KRAB A domain containing 1 FNDC1 0.021 fibronectin type III domain containing 1 HOXB2 0.021 homeobox B2 MRPS7 0.021 mitochondrial ribosomal protein S7 MAGI1 0.021 membrane associated guanylate kinase, WW and PDZ dom... OTUD1 0.016 OTU domain containing 1 TRPV2 0.016 transient receptor potential cation channel, subfami... LATS1 0.016 LATS homolog 1 SOHLH1 0.016 spermatogenesis and oogenesis specific basic helix-... SOHLH1 0.016 spermatogenesis and oogenesis specific basic helix-... GDF10 0.016 growth differentiation factor 10 precursor KIF24 0.016 kinesin family member 24 CHERP 0.016 calcium homeostasis endoplasmic reticulum protein [... ZNF687 0.016 zinc finger protein 687 LOC100128186 0.010 PREDICTED: hypothetical protein LOC100128186 0.010 PREDICTED: hypothetical protein LOC100128186 0.010 PREDICTED: hypothetical protein ADAMTS7 0.010 ADAM metallopeptidase with thrombospondin type 1 mot... SOX10 0.010 SRY (sex determining region Y)-box 10 RBM6 0.010 RNA binding motif protein 6 NACA 0.010 nascent polypeptide-associated complex alpha subuni... PLEKHA4 0.010 pleckstrin homology domain containing family A memb... ARHGAP6 0.010 Rho GTPase activating protein 6 isoform 4 ARHGAP6 0.010 Rho GTPase activating protein 6 isoform 1 TP53I13 0.010 tumor protein p53 inducible protein 13 COL22A1 0.010 collagen, type XXII, alpha 1 DDX54 0.010 DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 isoform 2 ... DDX54 0.010 DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 isoform 1...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
