
| Name: TNFSF9 | Sequence: fasta or formatted (254aa) | NCBI GI: 4507609 | |
|
Description: tumor necrosis factor (ligand) superfamily, member 9
|
Referenced in:
| ||
|
Composition:
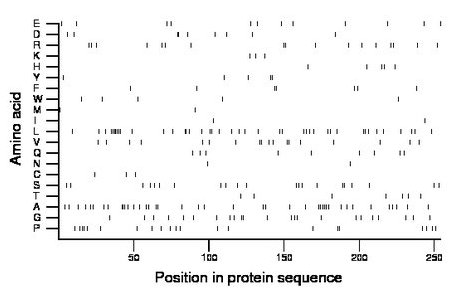
Amino acid Percentage Count Longest homopolymer A alanine 16.1 41 3 C cysteine 1.2 3 1 D aspartate 3.5 9 2 E glutamate 4.7 12 1 F phenylalanine 2.8 7 2 G glycine 9.1 23 2 H histidine 2.0 5 1 I isoleucine 0.8 2 1 K lysine 1.2 3 1 L leucine 16.5 42 7 M methionine 0.8 2 1 N asparagine 0.8 2 1 P proline 8.3 21 2 Q glutamine 3.5 9 1 R arginine 6.7 17 2 S serine 8.3 21 2 T threonine 2.8 7 1 V valine 7.1 18 2 W tryptophan 2.0 5 1 Y tyrosine 2.0 5 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 tumor necrosis factor (ligand) superfamily, member 9 ... TNFSF14 0.046 tumor necrosis factor ligand superfamily, member 14 ... TNFSF14 0.038 tumor necrosis factor ligand superfamily, member 14 ... MMP24 0.017 matrix metalloproteinase 24 preproprotein LOC100134152 0.013 PREDICTED: similar to synaptotagmin XV-b FOXF2 0.013 forkhead box F2 PLXNB1 0.013 plexin B1 PLXNB1 0.013 plexin B1 TNFSF12-TNFSF13 0.010 TNFSF12-TNFSF13 protein NPTXR 0.010 neuronal pentraxin receptor KCP 0.010 cysteine rich BMP regulator 2 isoform 2 KCP 0.010 cysteine rich BMP regulator 2 isoform 1 ETNK1 0.010 ethanolamine kinase 1 isoform B ETNK1 0.010 ethanolamine kinase 1 isoform A MSLNL 0.010 mesothelin-like TNFSF12 0.008 tumor necrosis factor (ligand) superfamily, member 12... LOC100292233 0.008 PREDICTED: hypothetical protein LOC100290077 0.008 PREDICTED: hypothetical protein XP_002347239 LOC100287086 0.008 PREDICTED: hypothetical protein XP_002343099 SOX21 0.008 SRY-box 21 LOC729991-MEF2B 0.006 myocyte enhancer factor 2B isoform b MAZ 0.006 MYC-associated zinc finger protein isoform 2 MAZ 0.006 MYC-associated zinc finger protein isoform 1 HDAC5 0.006 histone deacetylase 5 isoform 3 HDAC5 0.006 histone deacetylase 5 isoform 1 LOC100291961 0.006 PREDICTED: hypothetical protein XP_002345135 LOC100128328 0.006 PREDICTED: hypothetical protein LOC100128328 0.006 PREDICTED: hypothetical protein TAS1R3 0.006 taste receptor, type 1, member 3 SVIL 0.006 supervillin isoform 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
