
| Name: LOC100134274 | Sequence: fasta or formatted (148aa) | NCBI GI: 239755869 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
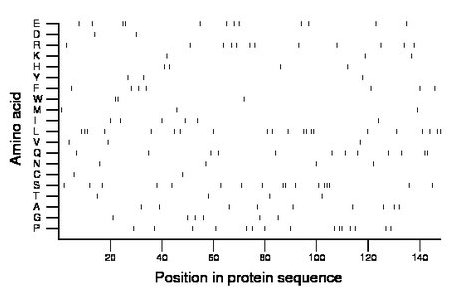
Amino acid Percentage Count Longest homopolymer A alanine 6.1 9 1 C cysteine 1.4 2 1 D aspartate 1.4 2 1 E glutamate 8.1 12 2 F phenylalanine 4.7 7 1 G glycine 4.1 6 1 H histidine 2.7 4 1 I isoleucine 4.1 6 1 K lysine 2.0 3 1 L leucine 14.2 21 3 M methionine 2.0 3 1 N asparagine 2.7 4 1 P proline 10.1 15 2 Q glutamine 8.1 12 2 R arginine 8.1 12 1 S serine 11.5 17 3 T threonine 2.7 4 1 V valine 2.0 3 1 W tryptophan 2.0 3 2 Y tyrosine 2.0 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein SEZ6L 0.036 seizure related 6 homolog (mouse)-like precursor [Ho... GGA1 0.029 golgi associated, gamma adaptin ear containing, ARF b... GGA1 0.029 golgi associated, gamma adaptin ear containing, ARF ... KIAA1543 0.029 NEZHA isoform 2 KIAA1543 0.029 NEZHA isoform 1 ZNF853 0.025 zinc finger protein 853 SSH3 0.025 slingshot homolog 3 C20orf151 0.025 hypothetical protein LOC140893 SYN1 0.022 synapsin I isoform Ia ZNF609 0.018 zinc finger protein 609 AFF4 0.018 ALL1 fused gene from 5q31 DACH1 0.018 dachshund homolog 1 isoform c LOC651986 0.018 PREDICTED: similar to forkhead box L2 NEDD4L 0.014 neural precursor cell expressed, developmentally do... NEDD4L 0.014 neural precursor cell expressed, developmentally dow... NEDD4L 0.014 neural precursor cell expressed, developmentally do... NEDD4L 0.014 neural precursor cell expressed, developmentally do... NEDD4L 0.014 neural precursor cell expressed, developmentally do... NEDD4L 0.014 neural precursor cell expressed, developmentally do... NEDD4L 0.014 neural precursor cell expressed, developmentally do... NEDD4L 0.014 neural precursor cell expressed, developmentally do... NEDD4L 0.014 neural precursor cell expressed, developmentally do... DACH1 0.014 dachshund homolog 1 isoform b DACH1 0.014 dachshund homolog 1 isoform a CITED2 0.014 Cbp/p300-interacting transactivator, with Glu/Asp-ri... ROBO4 0.014 roundabout homolog 4, magic roundabout LMTK3 0.014 lemur tyrosine kinase 3 TOX3 0.014 TOX high mobility group box family member 3 isoform... TOX3 0.014 TOX high mobility group box family member 3 isoform...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
