
| Name: LOC642587 | Sequence: fasta or formatted (102aa) | NCBI GI: 157151759 | |
|
Description: hypothetical protein LOC642587
| Not currently referenced in the text | ||
|
Composition:
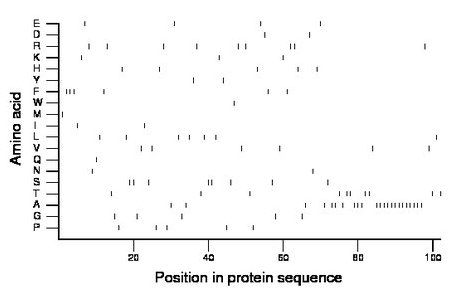
Amino acid Percentage Count Longest homopolymer A alanine 22.5 23 13 C cysteine 0.0 0 0 D aspartate 2.0 2 1 E glutamate 3.9 4 1 F phenylalanine 5.9 6 3 G glycine 4.9 5 1 H histidine 4.9 5 1 I isoleucine 2.0 2 1 K lysine 2.9 3 1 L leucine 6.9 7 1 M methionine 1.0 1 1 N asparagine 2.0 2 1 P proline 4.9 5 1 Q glutamine 1.0 1 1 R arginine 8.8 9 2 S serine 7.8 8 2 T threonine 9.8 10 2 V valine 5.9 6 1 W tryptophan 1.0 1 1 Y tyrosine 2.0 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC642587 ZSWIM6 0.085 zinc finger, SWIM-type containing 6 TMEM158 0.062 Ras-induced senescence 1 NKX6-1 0.056 NK6 transcription factor related, locus 1 ALKBH5 0.056 alkB, alkylation repair homolog 5 EVX2 0.045 even-skipped homeobox 2 FAM48B1 0.045 hypothetical protein LOC100130302 PRPF40A 0.045 formin binding protein 3 RCOR1 0.045 REST corepressor 1 RBM24 0.040 RNA binding motif protein 24 isoform 3 RBM24 0.040 RNA binding motif protein 24 isoform 2 RBM24 0.040 RNA binding motif protein 24 isoform 1 POU4F1 0.040 POU domain, class 4, transcription factor 1 SP9 0.040 Sp9 transcription factor homolog CHD3 0.040 chromodomain helicase DNA binding protein 3 isoform ... CHD3 0.040 chromodomain helicase DNA binding protein 3 isoform ... CHD3 0.040 chromodomain helicase DNA binding protein 3 isoform... LOC100287723 0.034 PREDICTED: hypothetical protein XP_002342271 DMRTA2 0.034 DMRT-like family A2 ZMIZ2 0.034 zinc finger, MIZ-type containing 2 isoform 1 ZMIZ2 0.034 zinc finger, MIZ-type containing 2 isoform 2 ZBTB8B 0.034 zinc finger and BTB domain containing 8B EN1 0.034 engrailed homeobox 1 PHOX2B 0.034 paired-like homeobox 2b GUCY1A2 0.034 guanylate cyclase 1, soluble, alpha 2 AMOT 0.034 angiomotin isoform 2 AMOT 0.034 angiomotin isoform 1 SOX21 0.034 SRY-box 21 HOXA13 0.028 homeobox A13 BTBD2 0.028 BTB (POZ) domain containing 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
