
| Name: PNRC2 | Sequence: fasta or formatted (139aa) | NCBI GI: 8923295 | |
|
Description: proline-rich nuclear receptor coactivator 2
|
Referenced in:
| ||
|
Composition:
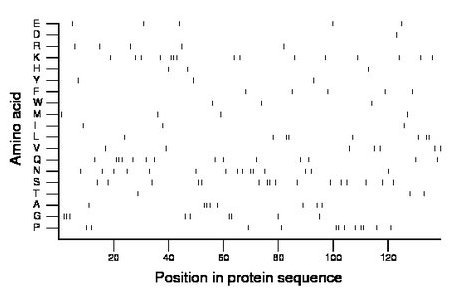
Amino acid Percentage Count Longest homopolymer A alanine 5.8 8 3 C cysteine 0.0 0 0 D aspartate 0.7 1 1 E glutamate 3.6 5 1 F phenylalanine 3.6 5 1 G glycine 6.5 9 3 H histidine 2.2 3 1 I isoleucine 2.2 3 1 K lysine 10.8 15 3 L leucine 5.8 8 2 M methionine 2.9 4 1 N asparagine 10.8 15 2 P proline 8.6 12 2 Q glutamine 10.1 14 3 R arginine 3.6 5 1 S serine 11.5 16 2 T threonine 2.2 3 1 V valine 5.0 7 1 W tryptophan 2.2 3 1 Y tyrosine 2.2 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 proline-rich nuclear receptor coactivator 2 LOC100131261 0.981 PREDICTED: similar to hCG1728885 LOC100131261 0.981 PREDICTED: similar to hCG1728885 LOC100131261 0.958 PREDICTED: hypothetical protein LOC100131261 PNRC1 0.231 proline-rich nuclear receptor coactivator 1 SMN2 0.034 survival of motor neuron 2, centromeric isoform d [H... SMN2 0.034 survival of motor neuron 2, centromeric isoform a [H... SMN1 0.034 survival of motor neuron 1, telomeric isoform d [Homo... LMOD2 0.034 leiomodin 2 (cardiac) GLIS3 0.030 GLIS family zinc finger 3 isoform a FBRSL1 0.030 fibrosin-like 1 GLIS3 0.030 GLIS family zinc finger 3 isoform b FGFR1OP2 0.027 FGFR1 oncogene partner 2 CHD9 0.019 chromodomain helicase DNA binding protein 9 N4BP2L2 0.019 phosphonoformate immuno-associated protein 5 isofor... C20orf151 0.019 hypothetical protein LOC140893 ZC3H18 0.019 zinc finger CCCH-type containing 18 PRPF38B 0.015 PRP38 pre-mRNA processing factor 38 (yeast) domain ... FOXO4 0.015 forkhead box O4 NKTR 0.015 natural killer-tumor recognition sequence TRDN 0.015 triadin C6orf222 0.015 hypothetical protein LOC389384 MINK1 0.011 misshapen/NIK-related kinase isoform 4 MINK1 0.011 misshapen/NIK-related kinase isoform 2 MINK1 0.011 misshapen/NIK-related kinase isoform 3 MINK1 0.011 misshapen/NIK-related kinase isoform 1 OBFC2B 0.011 oligonucleotide/oligosaccharide-binding fold contain... C14orf102 0.011 hypothetical protein LOC55051 isoform 1 MBTD1 0.011 mbt domain containing 1 FAM178A 0.011 hypothetical protein LOC55719 isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
