
| Name: C13orf35 | Sequence: fasta or formatted (121aa) | NCBI GI: 46409504 | |
|
Description: hypothetical protein LOC400165
| Not currently referenced in the text | ||
|
Composition:
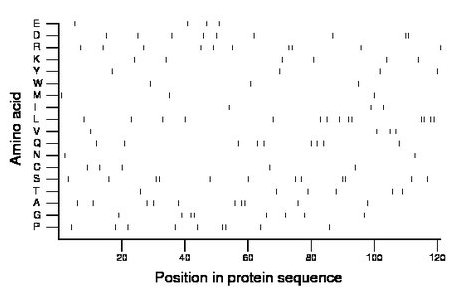
Amino acid Percentage Count Longest homopolymer A alanine 8.3 10 2 C cysteine 4.1 5 1 D aspartate 7.4 9 2 E glutamate 3.3 4 1 F phenylalanine 0.0 0 0 G glycine 6.6 8 2 H histidine 0.0 0 0 I isoleucine 2.5 3 1 K lysine 5.0 6 1 L leucine 11.6 14 2 M methionine 2.5 3 1 N asparagine 1.7 2 1 P proline 7.4 9 2 Q glutamine 7.4 9 1 R arginine 8.3 10 2 S serine 9.9 12 2 T threonine 5.0 6 1 V valine 3.3 4 1 W tryptophan 2.5 3 1 Y tyrosine 3.3 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC400165 PPP1R13L 0.026 protein phosphatase 1, regulatory subunit 13 like [... PPP1R13L 0.026 protein phosphatase 1, regulatory subunit 13 like [H... RAB11FIP3 0.018 rab11-family interacting protein 3 isoform 1 OIP5 0.013 Opa interacting protein 5 FLJ31485 0.013 PREDICTED: hypothetical protein C6orf47 0.013 G4 protein MGC50722 0.013 hypothetical protein LOC399693 DDN 0.009 dendrin CEP164 0.009 centrosomal protein 164kDa ZAR1L 0.009 zygote arrest 1-like LOC100291632 0.009 PREDICTED: hypothetical protein XP_002344527 LOC100288471 0.009 PREDICTED: hypothetical protein LOC100288471 0.009 PREDICTED: hypothetical protein XP_002343034 CCDC158 0.009 coiled-coil domain containing 158 PAK4 0.009 p21-activated kinase 4 isoform 1 PAK4 0.009 p21-activated kinase 4 isoform 1 PAK4 0.009 p21-activated kinase 4 isoform 1 ZCCHC4 0.009 zinc finger, CCHC domain containing 4 TMPRSS13 0.004 transmembrane protease, serine 13 LOC100287590 0.004 PREDICTED: hypothetical protein XP_002343592 LOC651856 0.004 PREDICTED: similar to hCG2023245Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
