
| Name: LOC100290466 | Sequence: fasta or formatted (68aa) | NCBI GI: 239749536 | |
|
Description: PREDICTED: hypothetical protein XP_002347073
| Not currently referenced in the text | ||
|
Composition:
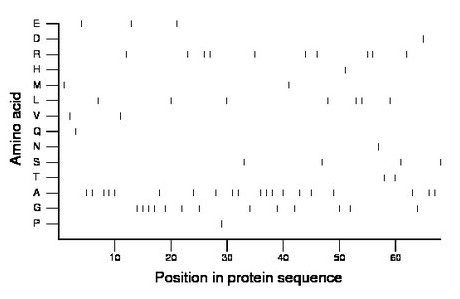
Amino acid Percentage Count Longest homopolymer A alanine 29.4 20 3 C cysteine 0.0 0 0 D aspartate 1.5 1 1 E glutamate 4.4 3 1 F phenylalanine 0.0 0 0 G glycine 19.1 13 4 H histidine 1.5 1 1 I isoleucine 0.0 0 0 K lysine 0.0 0 0 L leucine 10.3 7 2 M methionine 2.9 2 1 N asparagine 1.5 1 1 P proline 1.5 1 1 Q glutamine 1.5 1 1 R arginine 14.7 10 2 S serine 5.9 4 1 T threonine 2.9 2 1 V valine 2.9 2 1 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002347073 LOC100292904 1.000 PREDICTED: hypothetical protein LOC100289413 0.990 PREDICTED: hypothetical protein XP_002342934 LOC100129463 0.095 PREDICTED: hypothetical protein LOC100129463 0.095 PREDICTED: hypothetical protein LOC100129463 0.095 PREDICTED: hypothetical protein LOC100129463 0.095 PREDICTED: hypothetical protein CCDC85C 0.076 coiled-coil domain containing 85C LOC728767 0.076 PREDICTED: hypothetical protein LOC728767 0.076 PREDICTED: hypothetical protein MTMR1 0.076 myotubularin-related protein 1 HOXA13 0.057 homeobox A13 AVEN 0.057 cell death regulator aven SCRT1 0.057 scratch MLL 0.048 myeloid/lymphoid or mixed-lineage leukemia protein [... LOC100292370 0.048 PREDICTED: hypothetical protein LOC100292058 0.048 PREDICTED: hypothetical protein XP_002344766 LOC100290367 0.048 PREDICTED: hypothetical protein XP_002347417 LOC100288730 0.048 PREDICTED: hypothetical protein XP_002343287 WTIP 0.048 Wilms tumor 1 interacting protein LMO2 0.048 LIM domain only 2 isoform 1 USP31 0.048 ubiquitin specific peptidase 31 GDF7 0.048 growth differentiation factor 7 preproprotein ARID1B 0.048 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.048 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.048 AT rich interactive domain 1B (SWI1-like) isoform 3 ... LOC100287250 0.048 PREDICTED: hypothetical protein XP_002344493 ZSWIM6 0.048 zinc finger, SWIM-type containing 6 LOC100129036 0.038 PREDICTED: hypothetical protein FGF20 0.038 fibroblast growth factor 20Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
