
| Name: LOC100131271 | Sequence: fasta or formatted (95aa) | NCBI GI: 169207648 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {95aa} PREDICTED: hypothetical protein | |||
|
Composition:
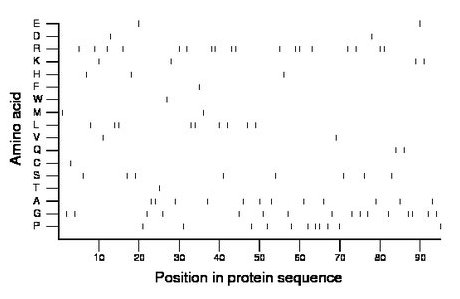
Amino acid Percentage Count Longest homopolymer A alanine 12.6 12 2 C cysteine 1.1 1 1 D aspartate 2.1 2 1 E glutamate 2.1 2 1 F phenylalanine 1.1 1 1 G glycine 16.8 16 2 H histidine 3.2 3 1 I isoleucine 0.0 0 0 K lysine 4.2 4 1 L leucine 9.5 9 2 M methionine 2.1 2 1 N asparagine 0.0 0 0 P proline 11.6 11 2 Q glutamine 2.1 2 1 R arginine 18.9 18 2 S serine 8.4 8 1 T threonine 1.1 1 1 V valine 2.1 2 1 W tryptophan 1.1 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100131271 1.000 PREDICTED: hypothetical protein LOC100289044 0.988 PREDICTED: hypothetical protein XP_002343344 LOC100132345 0.064 PREDICTED: hypothetical protein LOC100291129 0.058 PREDICTED: hypothetical protein XP_002347815 TESC 0.058 tescalcin SHANK3 0.058 SH3 and multiple ankyrin repeat domains 3 PPP1R10 0.052 protein phosphatase 1, regulatory subunit 10 ZCCHC2 0.052 zinc finger, CCHC domain containing 2 LRP10 0.052 low density lipoprotein receptor-related protein 10 ... TFPT 0.052 TCF3 (E2A) fusion partner C11orf35 0.052 hypothetical protein LOC256329 DUSP8 0.046 dual specificity phosphatase 8 SLC30A10 0.046 solute carrier family 30 (zinc transporter), member ... SMTNL2 0.046 smoothelin-like 2 isoform 1 BPTF 0.046 bromodomain PHD finger transcription factor isoform ... BPTF 0.046 bromodomain PHD finger transcription factor isoform ... SEC16B 0.040 leucine zipper transcription regulator 2 LOC100293513 0.040 PREDICTED: hypothetical protein LOC100294343 0.040 PREDICTED: hypothetical protein XP_002344111 FOXC1 0.040 forkhead box C1 LOC100128362 0.040 PREDICTED: similar to protein phosphatase 2, cataly... PRB1 0.040 proline-rich protein BstNI subfamily 1 isoform 1 pre... GATA4 0.035 GATA binding protein 4 LOC100127891 0.035 PREDICTED: hypothetical protein LOC645321 0.035 PREDICTED: hypothetical protein LOC100128074 0.035 PREDICTED: hypothetical protein LOC100291317 0.035 PREDICTED: hypothetical protein XP_002346735 LOC645321 0.035 PREDICTED: hypothetical protein COBL 0.035 cordon-bleu homologHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
