
| Name: MLANA | Sequence: fasta or formatted (118aa) | NCBI GI: 5031913 | |
|
Description: melan-A
|
Referenced in: Lysosomes and Related Organelles
| ||
|
Composition:
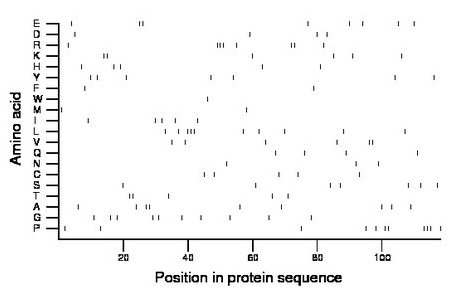
Amino acid Percentage Count Longest homopolymer A alanine 7.6 9 2 C cysteine 4.2 5 1 D aspartate 3.4 4 1 E glutamate 6.8 8 2 F phenylalanine 1.7 2 1 G glycine 8.5 10 1 H histidine 4.2 5 1 I isoleucine 4.2 5 1 K lysine 5.1 6 2 L leucine 8.5 10 3 M methionine 1.7 2 1 N asparagine 2.5 3 1 P proline 9.3 11 3 Q glutamine 3.4 4 1 R arginine 6.8 8 3 S serine 5.9 7 1 T threonine 4.2 5 2 V valine 5.1 6 2 W tryptophan 0.8 1 1 Y tyrosine 5.9 7 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 melan-A MLL2 0.031 myeloid/lymphoid or mixed-lineage leukemia 2 PILRB 0.026 paired immunoglobulin-like type 2 receptor beta isof... PILRB 0.026 paired immunoglobulin-like type 2 receptor beta isof... PCDH7 0.022 protocadherin 7 isoform b precursor PCDH7 0.022 protocadherin 7 isoform c precursor PCDH7 0.022 protocadherin 7 isoform a precursor ADAM19 0.017 ADAM metallopeptidase domain 19 preproprotein MTMR12 0.017 myotubularin related protein 12 INF2 0.013 inverted formin 2 isoform 2 INF2 0.013 inverted formin 2 isoform 1 C6orf132 0.009 PREDICTED: chromosome 6 open reading frame 132 [Hom... C6orf132 0.009 PREDICTED: chromosome 6 open reading frame 132 [Hom... C6orf132 0.009 PREDICTED: hypothetical protein LOC647024 DAB2IP 0.009 disabled homolog 2 interacting protein isoform 2 [Ho... DAB2IP 0.009 disabled homolog 2 interacting protein isoform 1 [Ho... ETV5 0.009 ets variant gene 5 (ets-related molecule) PHC1 0.009 polyhomeotic 1-like POLD3 0.009 DNA-directed DNA polymerase delta 3 SLC30A5 0.009 solute carrier family 30 (zinc transporter), member ... DRP2 0.009 dystrophin related protein 2 PTPRN2 0.009 protein tyrosine phosphatase, receptor type, N poly... PTPRN2 0.009 protein tyrosine phosphatase, receptor type, N poly... PTPRN2 0.009 protein tyrosine phosphatase, receptor type, N poly... FLJ22184 0.009 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.009 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.009 PREDICTED: hypothetical protein LOC80164 FAIM2 0.009 Fas apoptotic inhibitory molecule 2 PILRA 0.004 paired immunoglobulin-like type 2 receptor alpha iso... FAT1 0.004 FAT tumor suppressor 1 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
