
| Name: GPN1 | Sequence: fasta or formatted (279aa) | NCBI GI: 223005903 | |
|
Description: GPN-loop GTPase 1 isoform d
|
Referenced in: DNases, Recombination, and DNA Repair
| ||
Other entries for this name:
alt prot [388aa] GPN-loop GTPase 1 isoform a alt prot [362aa] GPN-loop GTPase 1 isoform b alt prot [295aa] GPN-loop GTPase 1 isoform c | |||
|
Composition:
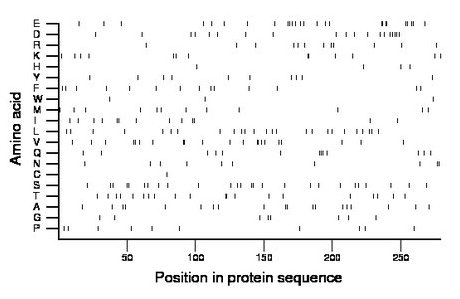
Amino acid Percentage Count Longest homopolymer A alanine 6.8 19 2 C cysteine 0.4 1 1 D aspartate 6.8 19 2 E glutamate 9.3 26 2 F phenylalanine 5.0 14 1 G glycine 2.9 8 1 H histidine 1.4 4 1 I isoleucine 4.7 13 2 K lysine 4.7 13 2 L leucine 9.0 25 1 M methionine 4.3 12 1 N asparagine 3.6 10 2 P proline 3.6 10 1 Q glutamine 4.7 13 3 R arginine 4.3 12 1 S serine 9.3 26 2 T threonine 7.2 20 2 V valine 7.5 21 2 W tryptophan 1.1 3 1 Y tyrosine 3.6 10 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 GPN-loop GTPase 1 isoform d GPN1 0.964 GPN-loop GTPase 1 isoform c GPN1 0.964 GPN-loop GTPase 1 isoform b GPN1 0.964 GPN-loop GTPase 1 isoform a GPN2 0.036 ATP binding domain 1 family, member B GPN3 0.030 GPN-loop GTPase 3 RP1L1 0.027 retinitis pigmentosa 1-like 1 GNL2 0.021 guanine nucleotide binding protein-like 2 (nucleolar)... GIGYF2 0.021 GRB10 interacting GYF protein 2 isoform c GIGYF2 0.021 GRB10 interacting GYF protein 2 isoform a GIGYF2 0.021 GRB10 interacting GYF protein 2 isoform b GIGYF2 0.021 GRB10 interacting GYF protein 2 isoform b MYH9 0.021 myosin, heavy polypeptide 9, non-muscle APOA4 0.015 apolipoprotein A-IV precursor TPR 0.015 nuclear pore complex-associated protein TPR SETD1B 0.015 SET domain containing 1B PBXIP1 0.015 pre-B-cell leukemia homeobox interacting protein 1 [... SPAG7 0.015 sperm associated antigen 7 UBTF 0.015 upstream binding transcription factor, RNA polymera... UBTF 0.015 upstream binding transcription factor, RNA polymera... UBTF 0.015 upstream binding transcription factor, RNA polymerase... MYST4 0.015 MYST histone acetyltransferase (monocytic leukemia)... ERMN 0.013 ermin, ERM-like protein isoform a ERMN 0.013 ermin, ERM-like protein isoform b NPM1 0.013 nucleophosmin 1 isoform 3 NPM1 0.013 nucleophosmin 1 isoform 2 NPM1 0.013 nucleophosmin 1 isoform 1 ERBB3 0.013 erbB-3 isoform 1 precursor CSMD3 0.011 CUB and Sushi multiple domains 3 isoform 2 CSMD3 0.011 CUB and Sushi multiple domains 3 isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
