
| Name: AMPD3 | Sequence: fasta or formatted (776aa) | NCBI GI: 4502079 | |
|
Description: adenosine monophosphate deaminase 3 isoform 1A
|
Referenced in: Purine Biosynthesis and Catabolism
| ||
Other entries for this name:
alt prot [767aa] adenosine monophosphate deaminase 3 isoform 1B alt prot [774aa] adenosine monophosphate deaminase 3 isoform 1C | |||
|
Composition:
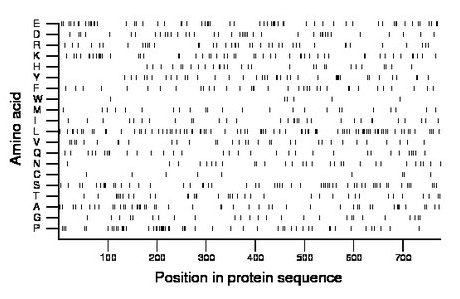
Amino acid Percentage Count Longest homopolymer A alanine 6.4 50 3 C cysteine 1.2 9 1 D aspartate 5.3 41 2 E glutamate 7.7 60 2 F phenylalanine 4.5 35 1 G glycine 3.6 28 2 H histidine 3.7 29 1 I isoleucine 4.0 31 2 K lysine 6.6 51 2 L leucine 10.8 84 3 M methionine 3.2 25 1 N asparagine 4.0 31 2 P proline 7.1 55 3 Q glutamine 4.3 33 2 R arginine 5.3 41 2 S serine 7.2 56 2 T threonine 4.8 37 2 V valine 4.3 33 2 W tryptophan 0.8 6 1 Y tyrosine 5.3 41 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 adenosine monophosphate deaminase 3 isoform 1A AMPD3 0.992 adenosine monophosphate deaminase 3 isoform 1C [Homo... AMPD3 0.990 adenosine monophosphate deaminase 3 isoform 1B [Homo... AMPD1 0.578 adenosine monophosphate deaminase 1 (isoform M) [Homo... AMPD2 0.486 adenosine monophosphate deaminase 2 (isoform L) isof... AMPD2 0.486 adenosine monophosphate deaminase 2 (isoform L) isof... AMPD2 0.480 adenosine monophosphate deaminase 2 (isoform L) isof... CECR1 0.013 cat eye syndrome critical region protein 1 isoform b... CECR1 0.013 cat eye syndrome critical region protein 1 isoform a... CCDC89 0.008 coiled-coil domain containing 89 SHROOM3 0.008 shroom family member 3 protein WHSC2 0.008 Wolf-Hirschhorn syndrome candidate 2 protein INPPL1 0.007 inositol polyphosphate phosphatase-like 1 PHF14 0.007 PHD finger protein 14 isoform 2 PHF14 0.007 PHD finger protein 14 isoform 1 ZNF207 0.006 zinc finger protein 207 isoform c ZNF207 0.006 zinc finger protein 207 isoform a ZNF207 0.006 zinc finger protein 207 isoform b CRKRS 0.006 Cdc2-related kinase, arginine/serine-rich isoform 2... CRKRS 0.006 Cdc2-related kinase, arginine/serine-rich isoform 1... TNRC6B 0.005 trinucleotide repeat containing 6B isoform 3 PPP4R4 0.005 HEAT-like repeat-containing protein isoform 1 BAZ2A 0.005 bromodomain adjacent to zinc finger domain, 2A [Homo... TAF3 0.005 RNA polymerase II transcription factor TAFII140 [Ho... ABI1 0.005 abl-interactor 1 isoform c DBF4B 0.005 DBF4 homolog B isoform 1 TSHZ3 0.005 zinc finger protein 537 BCL9L 0.005 B-cell CLL/lymphoma 9-like USP49 0.005 ubiquitin thioesterase 49 PXK 0.005 PX domain containing serine/threonine kinaseHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
