
| Name: LOC285141 | Sequence: fasta or formatted (327aa) | NCBI GI: 239753349 | |
|
Description: PREDICTED: hypothetical protein LOC285141
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {327aa} PREDICTED: hypothetical protein LOC285141 alt mRNA {327aa} PREDICTED: hypothetical protein LOC285141 | |||
|
Composition:
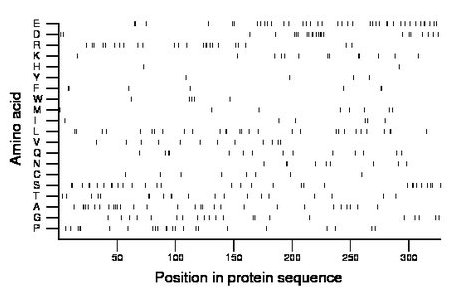
Amino acid Percentage Count Longest homopolymer A alanine 8.9 29 3 C cysteine 2.4 8 1 D aspartate 7.0 23 3 E glutamate 11.9 39 2 F phenylalanine 2.1 7 2 G glycine 6.4 21 2 H histidine 0.6 2 1 I isoleucine 1.8 6 1 K lysine 4.0 13 2 L leucine 8.9 29 2 M methionine 2.4 8 1 N asparagine 2.8 9 2 P proline 7.0 23 3 Q glutamine 4.3 14 2 R arginine 7.3 24 2 S serine 9.5 31 2 T threonine 6.1 20 2 V valine 3.7 12 1 W tryptophan 1.5 5 1 Y tyrosine 1.2 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC285141 LOC285141 1.000 PREDICTED: hypothetical protein LOC285141 LOC285141 1.000 PREDICTED: hypothetical protein LOC285141 HUWE1 0.030 HECT, UBA and WWE domain containing 1 MDN1 0.028 MDN1, midasin homolog DMP1 0.027 dentin matrix acidic phosphoprotein 1 isoform 1 precu... DMP1 0.027 dentin matrix acidic phosphoprotein 1 isoform 2 pre... EIF5B 0.025 eukaryotic translation initiation factor 5B NCL 0.024 nucleolin RGAG4 0.022 retrotransposon gag domain containing 4 MTERFD2 0.022 MTERF domain containing 2 TAF7L 0.022 TATA box binding protein-associated factor, RNA poly... ARID4A 0.020 retinoblastoma-binding protein 1 isoform III ARID4A 0.020 retinoblastoma-binding protein 1 isoform II ARID4A 0.020 retinoblastoma-binding protein 1 isoform I VPRBP 0.020 HIV-1 Vpr binding protein LOC100293445 0.020 PREDICTED: hypothetical protein LOC100288225 0.020 PREDICTED: hypothetical protein LOC100288225 0.020 PREDICTED: hypothetical protein XP_002343333 ATRX 0.020 transcriptional regulator ATRX isoform 2 ATRX 0.020 transcriptional regulator ATRX isoform 1 LOC647020 0.019 PREDICTED: similar to acidic (leucine-rich) nuclear... LOC647020 0.019 PREDICTED: similar to acidic (leucine-rich) nuclear... MYT1 0.019 myelin transcription factor 1 HTATSF1 0.019 HIV-1 Tat specific factor 1 HTATSF1 0.019 HIV-1 Tat specific factor 1 HIVEP2 0.019 human immunodeficiency virus type I enhancer bindin... CHD2 0.019 chromodomain helicase DNA binding protein 2 isoform... NSBP1 0.019 nucleosomal binding protein 1 AP3B1 0.019 adaptor-related protein complex 3, beta 1 subunit [H...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
