
| Name: LOC100289723 | Sequence: fasta or formatted (152aa) | NCBI GI: 239747205 | |
|
Description: PREDICTED: hypothetical protein XP_002346408
| Not currently referenced in the text | ||
|
Composition:
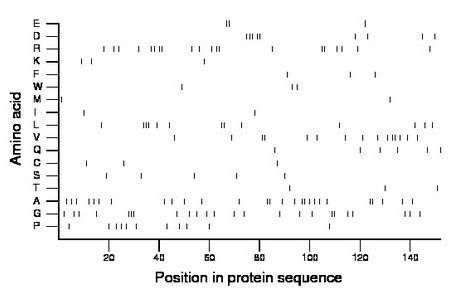
Amino acid Percentage Count Longest homopolymer A alanine 17.8 27 2 C cysteine 2.0 3 1 D aspartate 5.9 9 3 E glutamate 2.0 3 2 F phenylalanine 2.0 3 1 G glycine 15.8 24 3 H histidine 0.0 0 0 I isoleucine 1.3 2 1 K lysine 2.0 3 1 L leucine 8.6 13 3 M methionine 1.3 2 1 N asparagine 0.0 0 0 P proline 7.2 11 1 Q glutamine 3.9 6 1 R arginine 13.2 20 2 S serine 3.3 5 1 T threonine 2.0 3 1 V valine 9.9 15 2 W tryptophan 2.0 3 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346408 LOC100293554 0.543 PREDICTED: hypothetical protein LOC100288202 0.543 PREDICTED: hypothetical protein XP_002342060 ARTN 0.028 neurotrophic factor artemin isoform 2 ARTN 0.028 neurotrophic factor artemin isoform 1 precursor [Ho... ARTN 0.028 neurotrophic factor artemin isoform 3 precursor [Ho... ARTN 0.028 neurotrophic factor artemin isoform 1 precursor [Ho... ARTN 0.028 neurotrophic factor artemin isoform 3 precursor [Ho... LOC123855 0.028 PREDICTED: similar to hCG2042536 LOC123855 0.028 PREDICTED: similar to hCG2042536 LOC100134240 0.025 PREDICTED: hypothetical protein LOC339742 0.025 PREDICTED: hypothetical protein LOC100133636 0.025 PREDICTED: similar to hCG1806822 C9orf4 0.025 hypothetical protein LOC23732 LOC100292143 0.025 PREDICTED: hypothetical protein LOC100133885 0.025 PREDICTED: hypothetical protein LOC100133638 0.025 PREDICTED: hypothetical protein LOC100132261 0.025 PREDICTED: similar to hCG2023245 TCF23 0.025 transcription factor 23 SAMD11 0.025 sterile alpha motif domain containing 11 LOC339742 0.021 PREDICTED: hypothetical protein BSN 0.021 bassoon protein SRF 0.021 serum response factor (c-fos serum response element-b... LOC388079 0.021 PREDICTED: similar to hCG2042704 LOC727804 0.021 PREDICTED: hypothetical protein PRDM15 0.021 PR domain containing 15 isoform 1 LOC100134365 0.018 PREDICTED: similar to ankyrin repeat domain 36 [Hom... C17orf96 0.018 hypothetical protein LOC100170841 GNAS 0.018 GNAS complex locus XLas LOC651986 0.018 PREDICTED: similar to forkhead box L2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
