
| Name: LOC100134702 | Sequence: fasta or formatted (265aa) | NCBI GI: 239508698 | |
|
Description: PREDICTED: similar to mucin
| Not currently referenced in the text | ||
|
Composition:
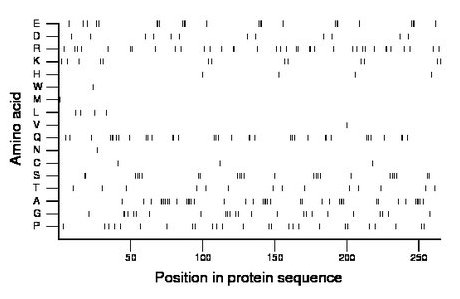
Amino acid Percentage Count Longest homopolymer A alanine 15.1 40 4 C cysteine 1.1 3 1 D aspartate 4.5 12 1 E glutamate 9.1 24 3 F phenylalanine 0.0 0 0 G glycine 9.1 24 2 H histidine 1.5 4 1 I isoleucine 0.0 0 0 K lysine 4.9 13 1 L leucine 1.5 4 1 M methionine 0.4 1 1 N asparagine 0.4 1 1 P proline 9.8 26 1 Q glutamine 13.6 36 3 R arginine 14.0 37 2 S serine 9.1 24 2 T threonine 5.3 14 1 V valine 0.4 1 1 W tryptophan 0.4 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to mucin LOC100293596 0.929 PREDICTED: similar to mucin LOC100288496 0.754 PREDICTED: hypothetical protein XP_002342747 LOC100292370 0.083 PREDICTED: hypothetical protein FLJ37078 0.067 hypothetical protein LOC222183 BASP1 0.063 brain abundant, membrane attached signal protein 1 [... APC2 0.061 adenomatosis polyposis coli 2 NOLC1 0.059 nucleolar and coiled-body phosphoprotein 1 SRRM2 0.059 splicing coactivator subunit SRm300 MARCKS 0.059 myristoylated alanine-rich protein kinase C substra... BPTF 0.059 bromodomain PHD finger transcription factor isoform ... RP1L1 0.057 retinitis pigmentosa 1-like 1 MAGI1 0.057 membrane associated guanylate kinase, WW and PDZ dom... CACNA1B 0.053 calcium channel, voltage-dependent, N type, alpha 1B ... MAP7D1 0.051 MAP7 domain containing 1 LOC100286959 0.051 PREDICTED: hypothetical protein XP_002343921 SRRM1 0.051 serine/arginine repetitive matrix 1 ANKRD56 0.051 ankyrin repeat domain 56 ARID1A 0.049 AT rich interactive domain 1A isoform a ARID1A 0.049 AT rich interactive domain 1A isoform b PRPF4B 0.049 serine/threonine-protein kinase PRP4K LOC643368 0.049 PREDICTED: similar to hCG1803335 ZC3H18 0.049 zinc finger CCCH-type containing 18 GPATCH8 0.049 G patch domain containing 8 FLG 0.047 filaggrin LOC100291634 0.047 PREDICTED: hypothetical protein XP_002346062 LOC100287232 0.047 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.047 PREDICTED: hypothetical protein PRB1 0.047 proline-rich protein BstNI subfamily 1 isoform 1 pre... BPTF 0.047 bromodomain PHD finger transcription factor isoform ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
