Comparative Genomics
Detection of homologs and assignment of functions
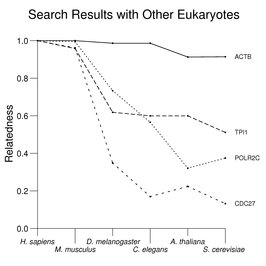
Searches for homologs often begin with a BLAST search against protein sequences from other species (see the Reference Protein Set for additional information about the search results used here). The following figure gives examples of such searches with four human proteins: ACTB (β actin), TPI1 (triose phosphate isomerase), POLR2C (an RNA polymerase II subunit), and CDC27 (an anaphase-promoting-complex subunit) against proteins from eukaryotic model systems (see About the Figures for more details about the scoring systems and limitations of the use of this method). Considerable variation in degree of sequence conservation is seen with these proteins even where related proteins in other species are relatively easy to identify.
The text contains many examples of the type shown above to illustrate various issues in comparative genomics. These include DNA replication proteins, cytoplasmic and mitochondrial aminoacyl tRNA synthetases, glycolytic enzymes, heat-shock proteins, G proteins, fragile X and associated functions, calcium signals, the Ras family, GPI-anchoring enzymes, the superoxide pathway, chloride channels, and membrane transporters.
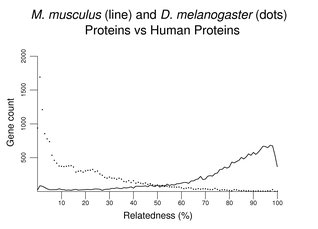
In the following figure, a largest protein isoform from each human gene (predictions excluded) was used in searches of mouse and D. melanogaster proteins. Scores (see About the Figures) were converted to % and rounded down (only identical proteins score 100), and grouped with a bin size of 1%. Relatively few human proteins have D. melanogaster matches of the degree typical with mouse proteins. A significant number of human proteins lack significant matches with D. melanogaster proteins.
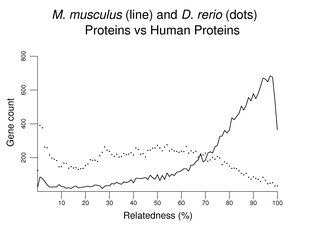
If a set of sea urchin proteins was used instead of a set of D. melanogaster proteins, the overall pattern would be similar to that above (not shown). The peak at the left would be somewhat smaller; the distribution would shift a bit to the right revealing many more matches at about 30% similarity. Another plot of this type follows, this time comparing mouse with zebrafish. Although the zebrafish distribution shifts far to the left, the peak at left remains relatively small compared to that with invertebrates (note the change in scale on the y-axis).
A basic test for assignment of function is determination that there is a reciprocal best match. One takes the best matching protein obtained in a BLAST search of another species and uses that protein in a reverse search of proteins from the original species. Failure to find the starting protein as the best match in the reverse search is not unusual and indicates that a simple homology relationship is not present.
Assignment of function becomes more challenging when the genes of interest are members of families in the species being studied. This occurs in highly conserved and in more diverged families. An interesting case where such complications arise involves C. elegans genes with differing ligand specificity and ion channel functions that are related to human GABA receptors. Other examples in the text involving S. cerevisiae genes include alcohol dehydrogenases, CDC20 family, CPSF subunits, the dual specificity protein phosphatase family, actin-related genes, the cyclins, and sequences related to a human RNA-editing enzyme.
Comparative genomics can provide hints for previously unsuspected biochemical pathways. One such case is fatty acid synthesis in mitochondria.
In some cases, particular steps in pathways may appear highly diverged or undetectable because of mechanistic or protein structure differences in the species being compared. For example, although the intermediates in glycolysis are the same in E. coli and human, their aldolases have unrelated sequences. Most enzymes that catalyze steps in the human histidine catabolic pathway can be identified from their Salmonella counterparts, but the glutamate formiminotransferase (see Amino Acid Catabolism) cannot.
Similarly, some of the S. cerevisiae pyrimidine biosynthetic genes readily identify their human counterparts, but S. cerevisiae encodes an unrelated dihydroorotase. Its dihydroorotate dehydrogenase uses a different mechanism and is also similar to a human pyrimidine catabolic enzyme.
Although bacteria and humans have related DNA cytosine 5-methyltransferases, the function of DNA methylation in bacterial cells is very different from that in mammals.
Although the counterparts of many human proteins are readily identified in diverse eukaryotes, with some human proteins one or more of the widely used model systems may not contain related sequences. Examples in the text include telomerase, the PARP (poly ADP-ribose polymerase) family, some lysosomal enzymes, and the pteridine cofactor. Yeast has proteins with ankyrin repeats but not clear counterparts of the ankyrins.
In the control of cell division, RB1-related proteins are readily identified in many species, but TP53-related proteins are not. A single TP53 family member can be detected by sequence similarity in D. melanogaster. C. elegans has a protein with TP53-like functions, but it is not readily detected by overall sequence similarity.
On occasion, a familiar protein will acquire a very different function during evolution. One well-known case is the function of a number of enzymes as crystallins in the eye of various vertebrates.
Relatives of human genes in many pathways and disease processes are often found in quite distant species. Some examples in the text include globin-like proteins, lysosomal diseases, adipocyte development, and otopetrin-related proteins.
Prokaryotic genomes
Despite the evolutionary distance, a considerable amount of information can be obtained through comparison of human and bacterial proteins. For highly conserved functions such as those of heat-shock proteins it is quite easy to identify bacterial counterparts. An interesting case is the relationship of human DNA polymerases to the well-studied E. coli enzymes.Although much can be learned from relationships to bacterial genes, several central components of human cells find counterparts in the archaea including parts of the transcriptional machinery (see RNA Polymerase and General Transcription Factors) and DNA replication proteins.
Additional comparisons of note involve nuclear-encoded mitochondrial functions and their similarity to prokaryotic proteins. Cytoplasmic translation factors find closer matches in archaea, whereas mitochondrial translation factors have closer relatives in bacteria. One interesting case is the relationship of mitochondrial RNA polymerase to bacteriophage RNA polymerases.
Bacterial sequences related to human genes are not confined to enzymes. Interesting examples are found with membrane proteins including potassium channels and aquaporins. See also the bacterial proteins related to the repeats in ankyrin.
Expansion of gene families
D. melanogaster contains counterparts to most of the MYC family. C. elegans has only a few genes of this type and none are readily identified by sequence similarity in S. cerevisiae. Both D. melanogaster and C. elegans have smaller E2F families and closely related proteins are not found in yeast.Many aspects of development were first explored in organisms such as D. melanogaster. A number of genes found as families in human are present as a single copy of D. melanogaster. Examples of this type include ephrin (and its receptor), hedgehog, and components of the notch pathway. Similar family expansions are seen relative to C. elegans. One case is the SLC34 group of phosphate solute carriers.
In the Wnt signaling pathway, both D. melanogaster and C. elegans have gene families for the ligands and receptors, but they are smaller than those seen in human. Similar situations are seen with the POU family of transcription factors and with the semaphorins (and their related receptors, the plexins). Although many components of the protein fucosylation pathways are single-copy genes in human, D. melanogaster, and C. elegans, one family of fucosyltransferases has expanded in human and another type found in humans lacks clear homologs in these two model systems.
The following table summarizes some of these data about gene family sizes based on the reference set data. Some metabolic enzymes are included for comparison. Because of widely dispersed repeated sequences, in some cases only a portion of the protein is suitable for family identification. Some predicted genes have been excluded. The C. elegans hedgehog proteins are quite different from those in the other two species.
| Comparison of Gene Family Sizes | |||
|---|---|---|---|
| Family | Gene counts | ||
| Human | D. melanogaster | C. elegans | |
| Alcohol dehydrogenase | 7 | 1 | 2 |
| Enolase | 3 | 1 | 1 |
| Nitric oxide synthase | 3 | 1 | 0 |
| E2F | 8 | 2 | 3 |
| Hedgehog | 3 | 1 | 10 |
| Notch (1401-1900) | 4 | 1 | 2 |
| Wnt | 19 | 7 | 5 |
| Ephrins | 8 | 1 | 4 |
| POU | 16 | 5 | 3 |
| SLC20 phosphate transporters | 2 | 1 | 6 |
| SLC34 phosphate transporters | 3 | 0 | 1 |
While S. cerevisiae has small gene families for components of the MAP kinase cascade, more proteins act at these steps in humans.
Family expansions are also seen in proteins involved in motor functions. Examples described in the text include myosins, tubulins, and kinesins. The spectrin family also provides clear examples of how mammals have evolved specialized functions not seen in the invertebrate model systems.
Protein interaction domain families in humans are often very large, involving proteins with diverse functions. These families can be much smaller in model systems—for example the yeast LIM domain proteins.
Although the human genome has a very large number of Ras-like small GTPases and associated proteins, this family expansion has not occurred in all branches of the family. Note the small number of Ran and associated proteins.
It is important to note that the human genome often contains smaller families than those seen in other species. One dramatic example is found with the olfactory receptors. Humans appear to encode many fewer functional members of the main OR family and it is not clear which, if any, of the few remaining vomeronasal receptor genes are functional. Humans also have fewer function type 2 (bitter) taste receptors. Many pseudogenes in these families are also present, complicating the determination of exact family sizes.
| Olfactory and Taste Receptors | ||
|---|---|---|
| Family | Gene counts | |
| Human | Mouse | |
| Olfactory | ~375 | ~1100 |
| Vomeronasal 1 | 0–5? | ~150 |
| Taste 2 | ~25 | ~33 |
Another example with larger gene families in other species is seen with the aromatic amino acid decarboxylases. This small family is larger in both D. melanogaster and C. elegans than in human.
When smaller human gene families are compared to those of other mammals, conservation of gene family structure is quite high but considerable variation exists. One case described in the text involves the serotonin receptors.
Many human oncogenes were identified as the counterparts of transforming genes discovered with avian or murine retroviruses. A number of these are present in mammalian genomes as large families (see, e.g., the Ras-like proteins). A large fraction of the human genome consists of sequences related to mobile elements of diverse species. A few of these transposon-related sequences have been suggested to have specific functions.
Notes and references
Many references and other information for individual genes can be found in the RefSeq entries linked via the pages for the proteins mentioned in this section. A table of these entries (with the corresponding gene identifiers) and a collection of their sequences also are available.Search results were obtained with NCBI BLASTP 2.2.11 and RefSeq proteins.
See also the additional reading for this chapter.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.