
| Name: MGAT1 | Sequence: fasta or formatted (445aa) | NCBI GI: 167857784 | |
|
Description: mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
Referenced in:
| ||
Other entries for this name:
alt mRNA [445aa] mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminylt... alt mRNA [445aa] mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminylt... alt mRNA [445aa] mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminylt... alt mRNA [445aa] mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminylt... | |||
|
Composition:
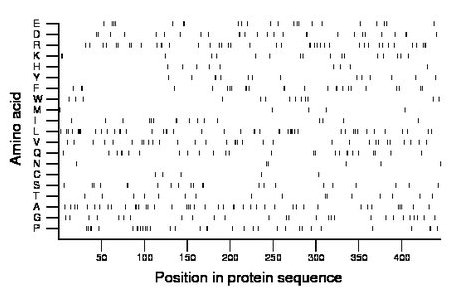
Amino acid Percentage Count Longest homopolymer A alanine 9.7 43 2 C cysteine 1.1 5 1 D aspartate 6.1 27 2 E glutamate 4.9 22 2 F phenylalanine 5.6 25 2 G glycine 6.1 27 1 H histidine 2.0 9 1 I isoleucine 3.6 16 2 K lysine 3.8 17 2 L leucine 10.3 46 4 M methionine 1.1 5 1 N asparagine 1.3 6 1 P proline 7.4 33 2 Q glutamine 5.8 26 2 R arginine 9.2 41 2 S serine 4.9 22 2 T threonine 3.6 16 1 V valine 7.2 32 3 W tryptophan 2.9 13 1 Y tyrosine 3.1 14 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... MGAT1 1.000 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... MGAT1 1.000 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... MGAT1 1.000 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... MGAT1 1.000 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... POMGNT1 0.127 O-linked mannose beta1,2-N-acetylglucosaminyltransf... MELK 0.011 maternal embryonic leucine zipper kinase CD1D 0.008 CD1D antigen precursor RAD52 0.008 RAD52 homolog MNT 0.008 MAX binding protein CMPK2 0.007 UMP-CMP kinase 2 C6orf132 0.006 PREDICTED: chromosome 6 open reading frame 132 [Hom... LOC730254 0.006 PREDICTED: hypothetical protein C6orf132 0.006 PREDICTED: chromosome 6 open reading frame 132 [Hom... C6orf132 0.006 PREDICTED: hypothetical protein LOC647024 LOC730254 0.006 PREDICTED: hypothetical protein C19orf61 0.006 hypothetical protein LOC56006 GLTSCR1 0.006 glioma tumor suppressor candidate region gene 1 [Ho... PALM2-AKAP2 0.006 PALM2-AKAP2 protein isoform 2 PALM2-AKAP2 0.006 PALM2-AKAP2 protein isoform 1 AKAP2 0.006 A kinase (PRKA) anchor protein 2 isoform 2 AKAP2 0.006 A kinase (PRKA) anchor protein 2 isoform 1 LOC100131908 0.006 PREDICTED: similar to forkhead box O6 FOXO6 0.006 PREDICTED: forkhead box protein O6 HEG1 0.006 HEG homolog 1 DMBT1 0.006 deleted in malignant brain tumors 1 isoform c precu... DMBT1 0.006 deleted in malignant brain tumors 1 isoform b precu... FYCO1 0.006 FYVE and coiled-coil domain containing 1 KIAA1522 0.006 hypothetical protein LOC57648 LOC100292259 0.004 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
