
| Name: PI4K2A | Sequence: fasta or formatted (479aa) | NCBI GI: 13559514 | |
|
Description: phosphatidylinositol 4-kinase type 2 alpha
|
Referenced in:
| ||
|
Composition:
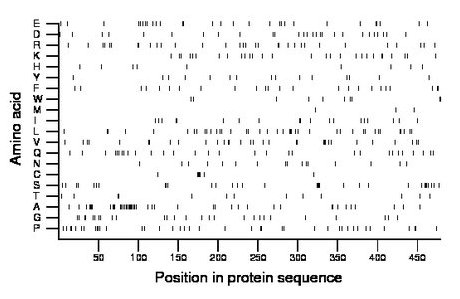
Amino acid Percentage Count Longest homopolymer A alanine 9.4 45 5 C cysteine 1.5 7 2 D aspartate 5.6 27 1 E glutamate 6.1 29 2 F phenylalanine 5.0 24 2 G glycine 5.6 27 2 H histidine 1.9 9 1 I isoleucine 4.0 19 2 K lysine 5.8 28 2 L leucine 8.4 40 3 M methionine 0.8 4 1 N asparagine 3.3 16 1 P proline 8.4 40 2 Q glutamine 6.3 30 1 R arginine 6.7 32 2 S serine 6.7 32 4 T threonine 2.7 13 1 V valine 6.7 32 3 W tryptophan 2.1 10 2 Y tyrosine 3.1 15 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 phosphatidylinositol 4-kinase type 2 alpha PI4K2B 0.557 phosphatidylinositol 4-kinase type 2 beta SOX12 0.015 SRY (sex determining region Y)-box 12 PHOX2B 0.013 paired-like homeobox 2b LOC728650 0.011 PREDICTED: hypothetical protein MLL2 0.010 myeloid/lymphoid or mixed-lineage leukemia 2 NR2F2 0.010 nuclear receptor subfamily 2, group F, member 2 isof... LOC728650 0.010 PREDICTED: hypothetical protein ZIC2 0.010 zinc finger protein of the cerebellum 2 NLK 0.010 nemo like kinase HECA 0.010 headcase SHROOM1 0.009 shroom family member 1 ARID3A 0.008 AT rich interactive domain 3A (BRIGHT- like) protein ... CASKIN1 0.008 CASK interacting protein 1 EPB41L3 0.008 erythrocyte membrane protein band 4.1-like 3 NKX2-4 0.008 NK2 homeobox 4 DDX51 0.008 DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 NFKBIE 0.008 nuclear factor of kappa light polypeptide gene enhan... LOC100133636 0.008 PREDICTED: similar to hCG1806822 LOC100288573 0.008 PREDICTED: hypothetical protein XP_002343799 TSC22D2 0.008 TSC22 domain family, member 2 BTBD2 0.008 BTB (POZ) domain containing 2 SNIP 0.008 SNAP25-interacting protein MEX3C 0.008 ring finger and KH domain containing 2 BTBD11 0.008 BTB (POZ) domain containing 11 isoform a AHDC1 0.008 AT hook, DNA binding motif, containing 1 LOC100292866 0.007 PREDICTED: hypothetical protein LOC100290853 0.007 PREDICTED: hypothetical protein XP_002348232 LOC100288457 0.007 PREDICTED: hypothetical protein XP_002342255 GRIN2D 0.007 N-methyl-D-aspartate receptor subunit 2D precursor ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
